WARNING WARNING WARNING
Nothing in this article should be considered medical advice or be diagnostic in any way. This article is purely for educational purposes for those interested in learning about the basic concepts that underlie genetic testing.
The bad news and some background on genetic diseases
Due to a recent harrowing and ongoing medical odyssey in my immediate family I learned that I have a 50% change of having autosomal dominant polycystic kidney disease.
Basically, there is a 50% change I inherited a copy of the polycistic kidney disease II gene that contains a missense mutation. This type of mutation changes a single base pair (letter) of the DNA sequence. DNA is the “recipe” for proteins which are the molecules that are actually doing work within your cells. Proteins themselves are composed of chains of amino acids. The specific DNA sequences within the coding regions of your genes determines the identity and order of these amino acids which ultimately gives rise to a protein’s function.
In my potential case, the change in one letter of the DNA recipe is enough to change the identify of one important amino acid within the PKD2 protein. Since the disease displays a dominant pattern of inheritance you only need to inherit one copy of the mutated gene (either from your Mom or your Dad) to be affected. Sticking with the recipe analogy for DNA, each person effectively carries two independent, largely identical, recipe books within each of their cells. Both these books are being used simultaneously by the cell to produce proteins. Therefore, one book might contain an error while the other does not and so about half the proteins you produce under this situation will be mutated. In many cases, having half the dose of normal protein is good enough for normal function. This is characteristic of diseases with autosomal recessives patterns of inheritance.
In diseases with autosomal dominant inheritance patterns you often need two fully functional copies of this gene to get the correct dose of protein for normal function or mutating one copy of the gene somehow actively interferes with normal physiological processes.
I won’t know my status as effected or unaffected without genetic testing. So being a genetics PhD student at home over winter break I decided to go through the process of designing my own genetic test that I could (in theory) carry out and then detail the process for educational purposes.
Test protocol outline
In order to do this test ourselves we will somehow need to determine the base pair that is present in both copies of my DNA at the site of the mutation. Since we only need to do this for one gene we can use some methodology that would have been available since the early 2000s which would in theory make this method very cheap.
- Identify the location of the particular PKD2 mutation that I may have inherited within the human genome
- Design primers for a PCR (Polymerase chain reaction) that will amplify the DNA region surrounding the site of the mutation. PCR allows us to create a lot of copies of a specific region of DNA that is defined by the primers we pick. The sequencing method we will use requires a large number of copies of a specific region in order to work well which motivates out choice to use PCR. PCR is a central technique in molecular biology, you can read more about it at this link.
This is where design ends and remaining steps would have to be actually carried out in the lab and so I won’t detail them here.
- Order PCR primers (this is very easy and cheap to do online)
- Extract my genomic DNA from a blood sample (finger prick)
- Amplify this extracted DNA using the PCR primers designed in step 2
- Purify the PCR product away from any remaining genomic DNA
- Send purified DNA for Sanger sequencing. A highly accurate and cheap sequencing method.
Locating the mutation
Using the report shared by our genetic counselor, I first
identified the location of the missense mutation in question
within the human genome. The particular mutation identified
is located in chromosome 4 at base pair 88,038,372 so
I pasted these coordinates into the UC Santa Cruz genome
browser Get DNA Window utility, making sure I used the same genome assembly as was listed in the diagnostic report.
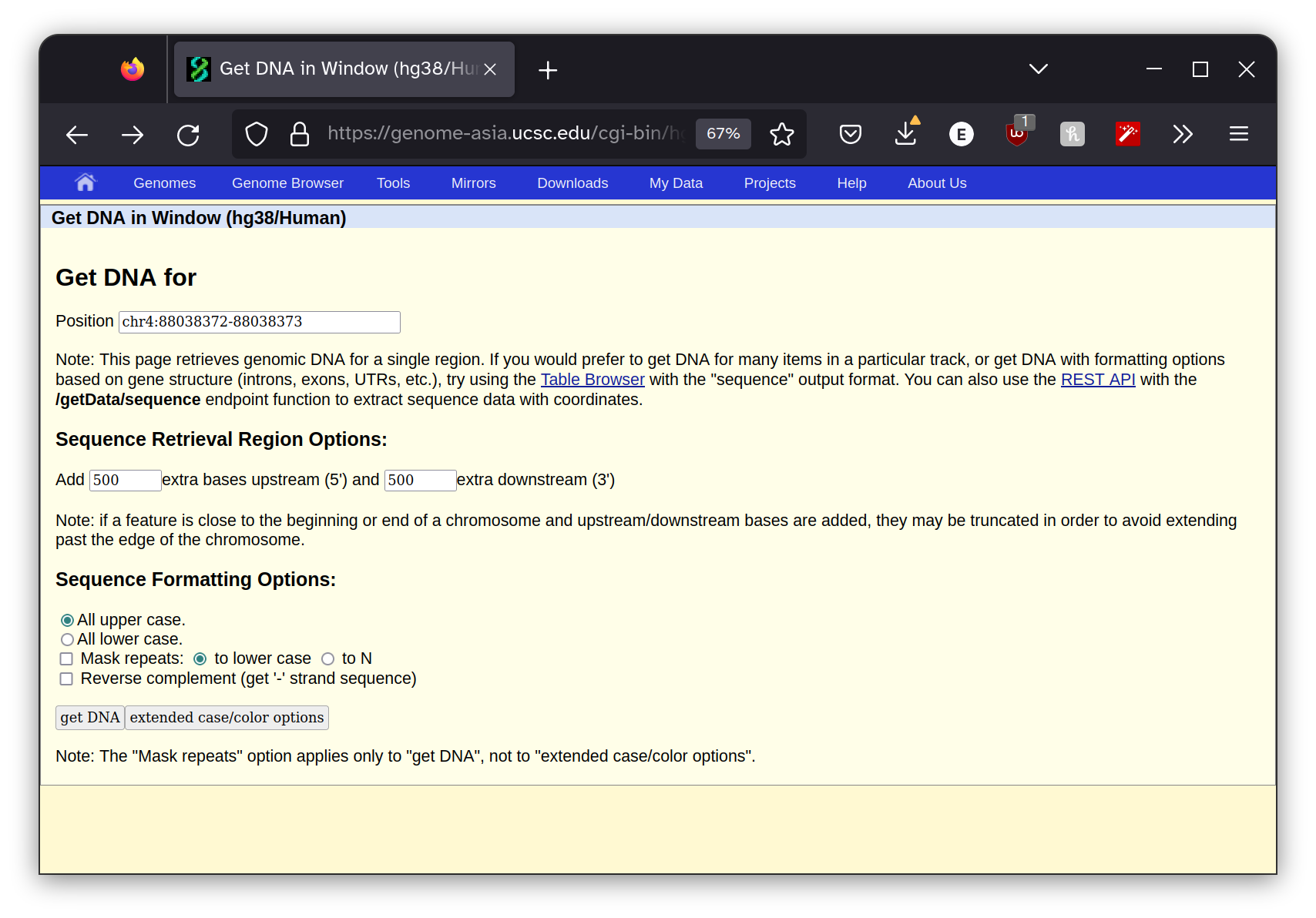
I also instructed the program to return 500 bp to the right and left of the site surrounding the mutation. This gives us the context that the mutation is occurring within which will be useful in the next step. The program returned this sequence.
>hg38_dna range=chr4:88037872-88038873 5'pad=500 3'pad=500 strand=+ repeatMasking=none
CAAAGTTTAGAACCATGATTGTGTGGGACATTATTTTAATAAGGGAAAGT
GCAGTTAATCATGACCCCACCTTTAGTCCAAGAACAAAAATCAGAGCTGC
CACGTATTAAGTACCCACTCTGTGCCAGGTGCAGTAACTATGCAAAAGAT
GGGTTTTCCAGATGCAAGAACCTTGGTTCAGAGGACCCTGCTCAAGGCCT
CATAGCTAACAAATGATGGGGCAAGATGCTATCCCAAATCTCTCTGACAA
CAAAACTCATTCTTATCACTCTACTATTTTCATAGAGTTGCCAAATGCTT
GGTTATGCAAACGATGCAGGCAGGGGCAAGACAGCGGCTGAGCTTGGAAC
TTTTTCAGAGATGTTTCCTTTGCTTTTAGTTCACAGAAGGCTCCTTATTG
GATGGGCTGTACTGGAAGATGCAGCCCAGCAACCAGACTGAAGCTGACAA
CCGAAGTTTCATCTTCTATGAGAACCTGCTGTTAGGGGTTCCACGAATAC
GGCAACTCCGAGTCAGAAATGGATCCTGCTCTATCCCCCAGGACTTGAGA
GATGAAATTAAAGAGTGCTATGATGTCTACTCTGTCAGTAGTGAAGATAG
GGCTCCCTTTGGGCCCCGAAATGGAACCGCGTAAGTGTCTGTGACTCATT
GCCACTCGGTGATATTCATTCATTTATTCTCTGAACTCCCACCATTCATT
CATTCATTCCCTGACACCTTCACCAAGGCAAAAATAAGTTCAGTGACTCT
TCAGTGCTTATATTTAAACCTTGGCCAACTTGACCTTTGACTTCTTAAGT
TTTCACTACTTCTTAGCCTTCTTTTAGTTTCTACATGCATATTTTTCAGA
AGACTAAATCGTTGACCATATAACCCCTCAAAAATTAATTATCTGAGCGT
TTGAAAATTTCATTTAAGATGCCCTGGGCCCTGTTTTTACAGGTGCAGTA
ACATCATCCACTAAGTTATTTAACACAAGTTTTCTGGTTCAGGAACTCTT
TT
Designing PCR primers
Next I wanted to create primers that would amplify a 300-600 bp region that included the site of the mutation. PCR uses two primers, a forward and reverse, to define the amplified region. The DNA that is between the forward and reverse primer will be amplified and become the target PCR product.
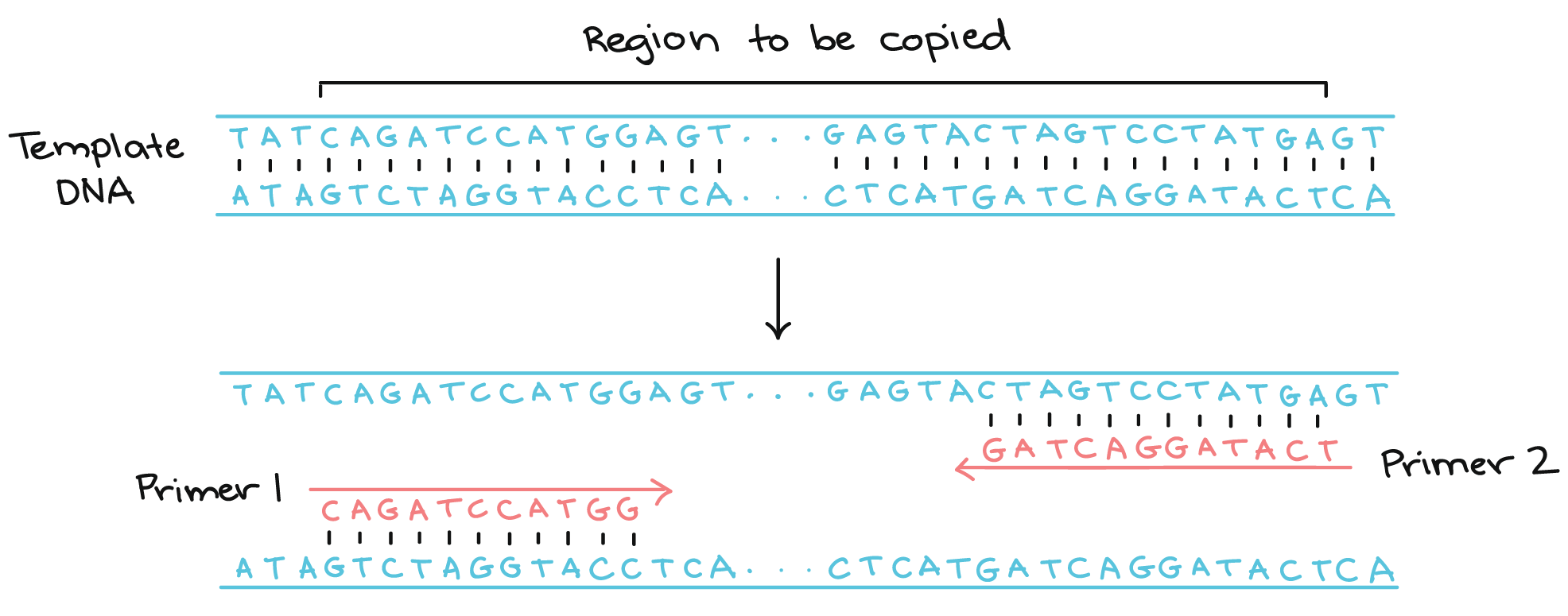
PCR diagram courtesy of Khan Academy
There are a large number of factors that can effect how well a PCR reaction works and picking good primers is a significant part of this calculus. So it is best to leave this decision up to well-established algorithms that can optimize primers for your specific sequence and product requirements. I prefer Primer3 so I entered my sequence into their interface and specified that I wanted 400-600 bp between the forward and reverse primers and that the region between the two primers must include the site of the PKD2 mutation I may have inherited.
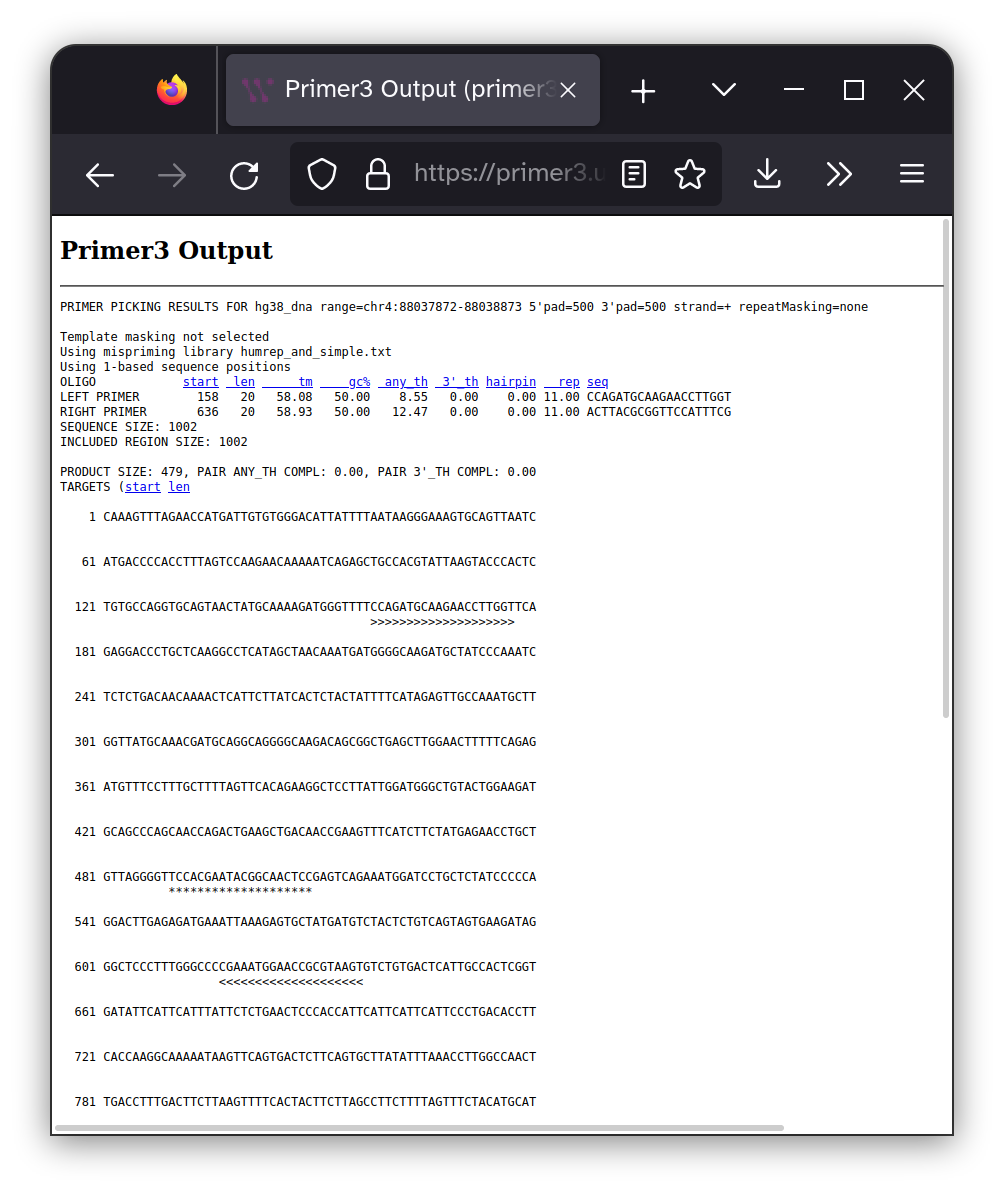
The location of the of each of the primers are shown within
the context of the target DNA sequence as >>>>>>>>>>>
characters and the 20 bp surrounding the mutation site are highlighted with ************** characters. The DNA sequence
of the primers are shown below.
Forward primer: CCAGATGCAAGAACCTTGGT
Reverse primer: ACTTACGCGGTTCCATTTCG
I also checked to make sure these primers would be specific to the region of the genome we are interested in using UCSC in silico PCR tool. This tool helps ensure your primers will only amplify your target region.
The next step would to be order these primers from a custom DNA oligo supplier like IDT.
Heading into the lab
With all our materials prepared we are ready to head into the lab and start working. First, using a sample of my blood I would extract the DNA. Many companies sell kits to make DNA extraction from whole blood easy, however these kits should not be used for diagnostic purposes.
After DNA is extracted from the blood we can use it as the template in a standard PCR reaction. If you are interested in learning more about this process AddGene as a great detailed article on the subject.
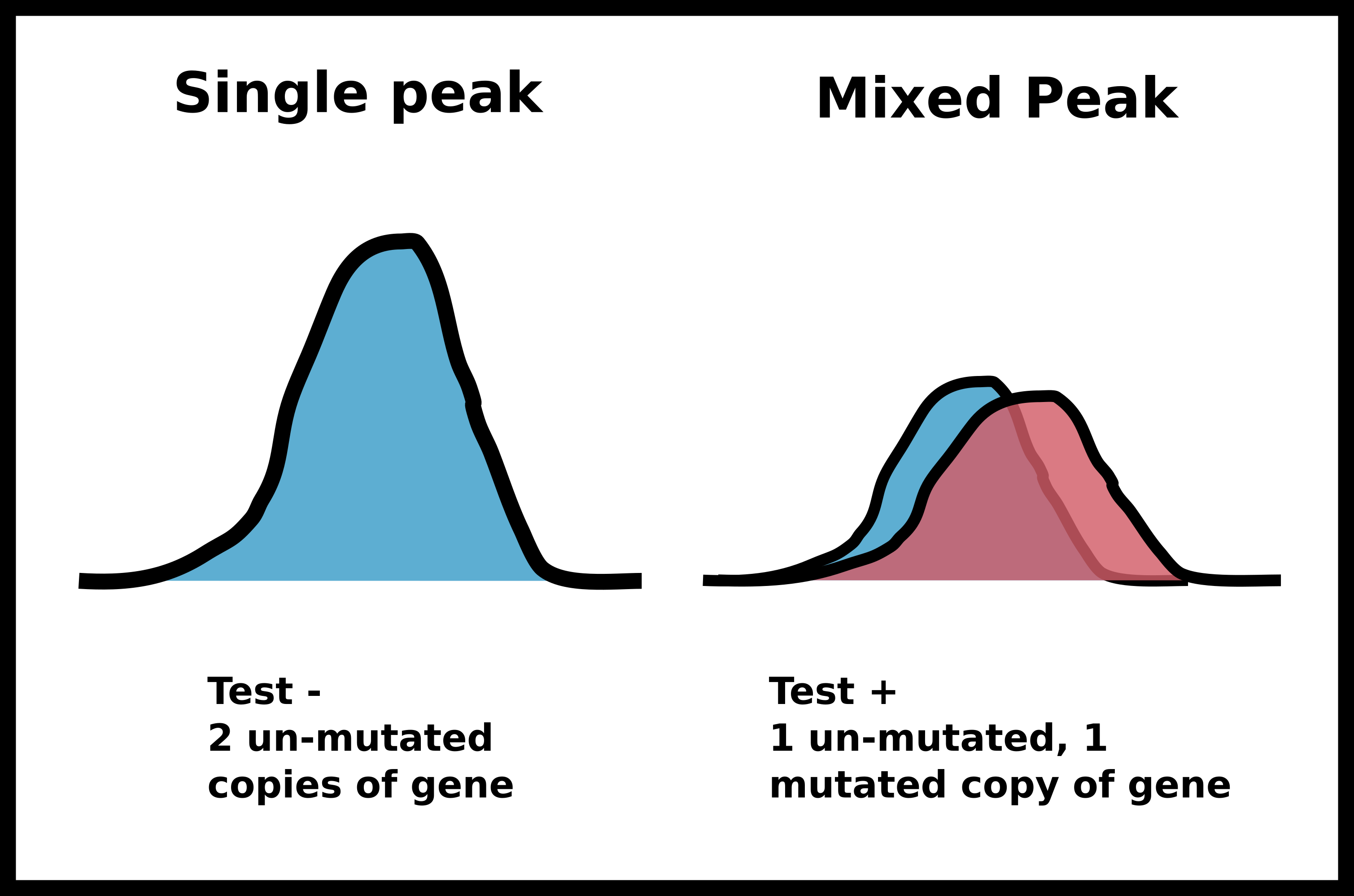
After this PCR reaction is completed we will send the amplified DNA out for sanger sequencing. Sanger sequencing is an analog method that produces “peaks” of signal corresponding to a specific base pair. If the identity of a base pair within our PCR product is the same within all DNA molecules in our sample the peak at that location will be very strong. However, if there is a mix of base pairs at the position multiple peaks will be present at that position. We can utilize this analog information to infer if a person has zero, one or two copies of a specific mutation such as the PKD2 missense mutation I may have inherited.
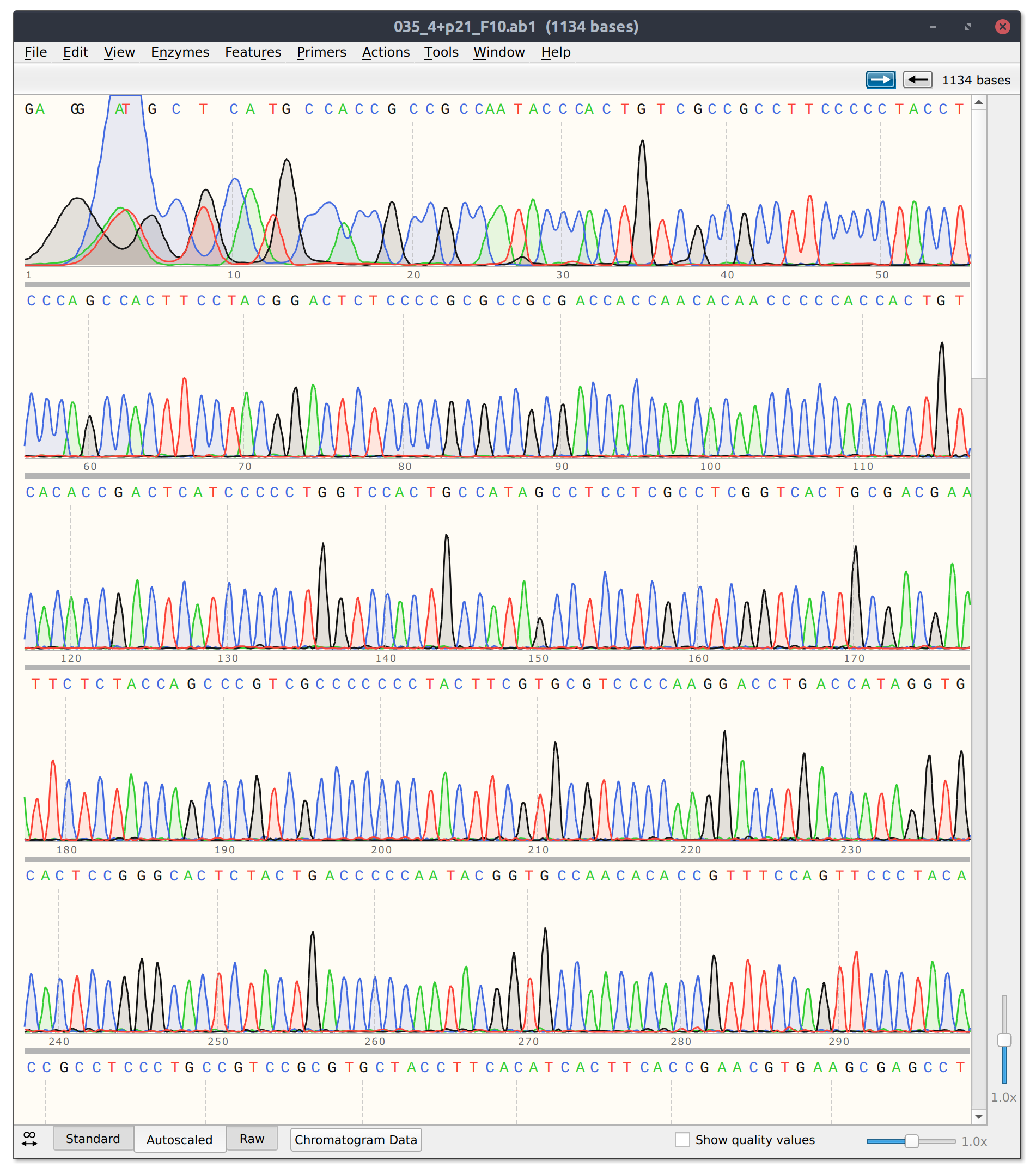
Example of sanger sequencing data from my own research
If in this example I did inherit the mutation (meaning one of two copies of my DNA has the single base pair change) I would expect the sanger sequencing peak to be mixed. If I did not have the mutation then the signal should be clean and monolithic.
I hope this article has helped you learn something about genetics and some of the experimental methodology behind the interesting questions we might want to ask. I again want to point out that this article is for educational purposes only and non of the materials described here should actually be used for medical diagnostics. Additionally, my approach described here is far from cutting edge. If you want the nitty-gritty of how genetic testing actually works in the clinic head straight to the source and check out the resources listed in Next-Generation Sequencing Resources for Pathologists from Illumina.