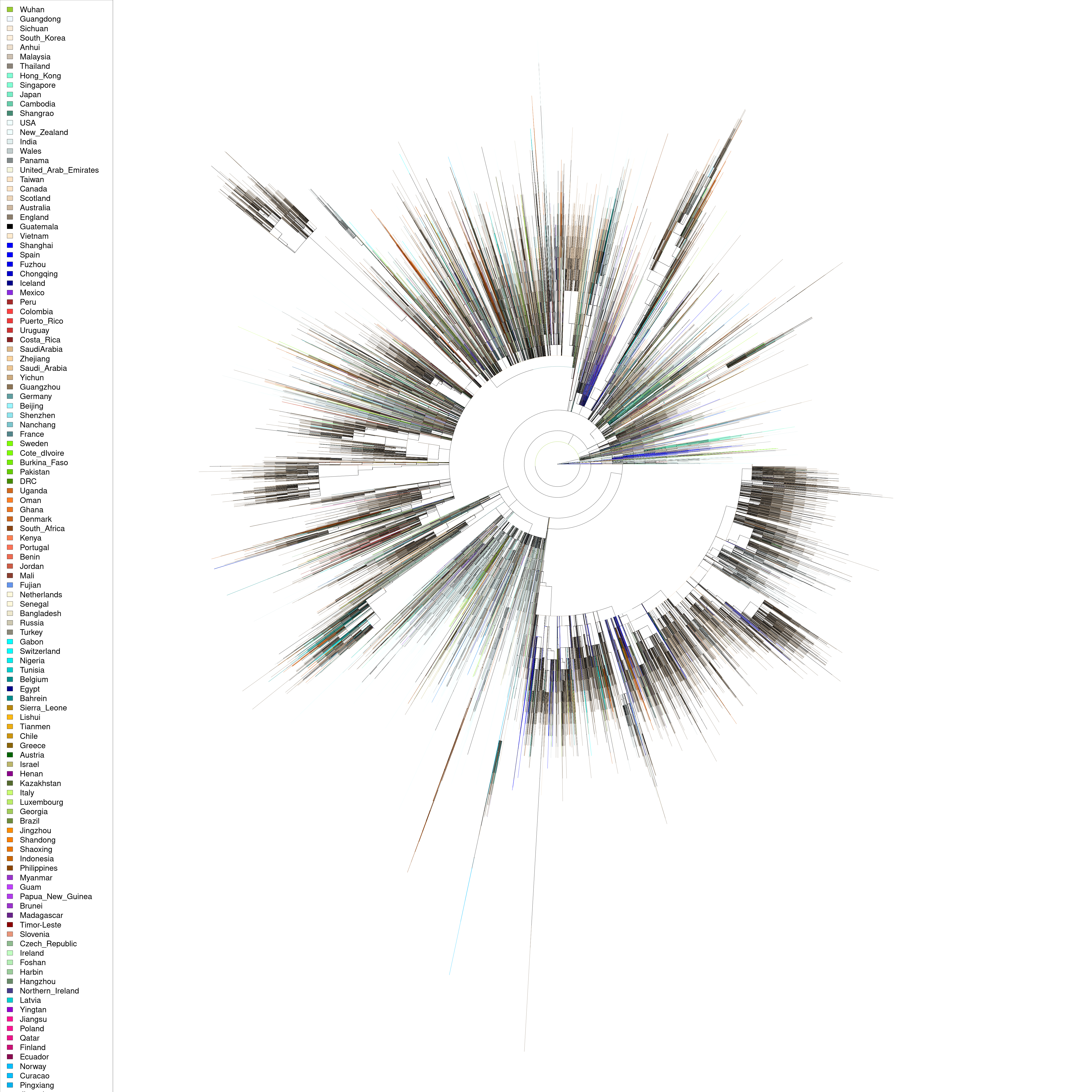
The Covid-19 Genomics UK Consortium has been collecting and sequencing thousands of COVID-19 genomes from patients in the UK and around the world.
All of their data is publicly available. Here I played around with the phylogenetic tree they have created from global alignments of all the genomes they have sequenced.
You can download the tree in Newick format from their data page which also hosts sequences and the alignment files.
Visualizing the COVID-19 phylogenetic tree by country of origin
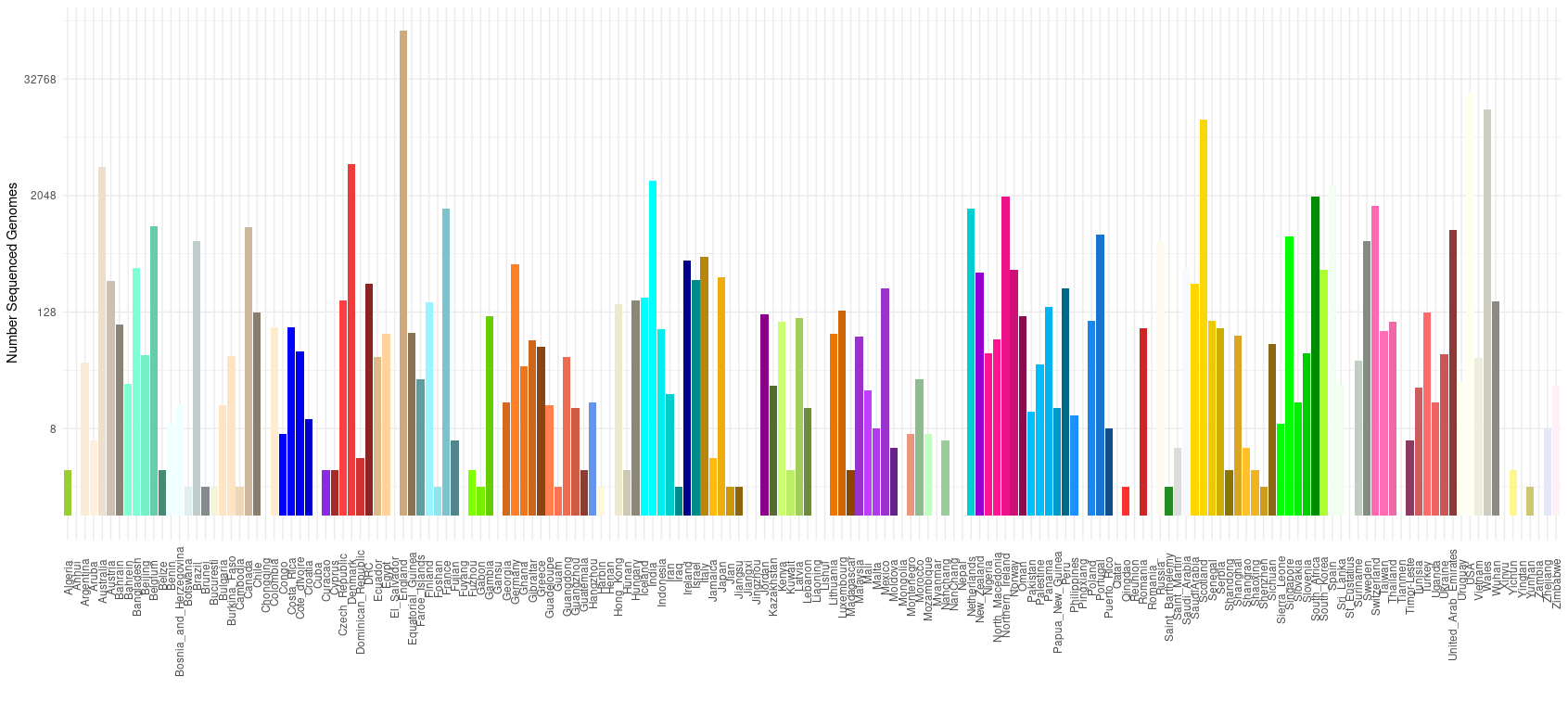
Genome count by country
Note this plot is log scale in the y-axis.
16 most prevalent UK COVID-19 lineages
Density plots showing the number of genomes of the 16 most prevalent lineages detected by COG-UK.
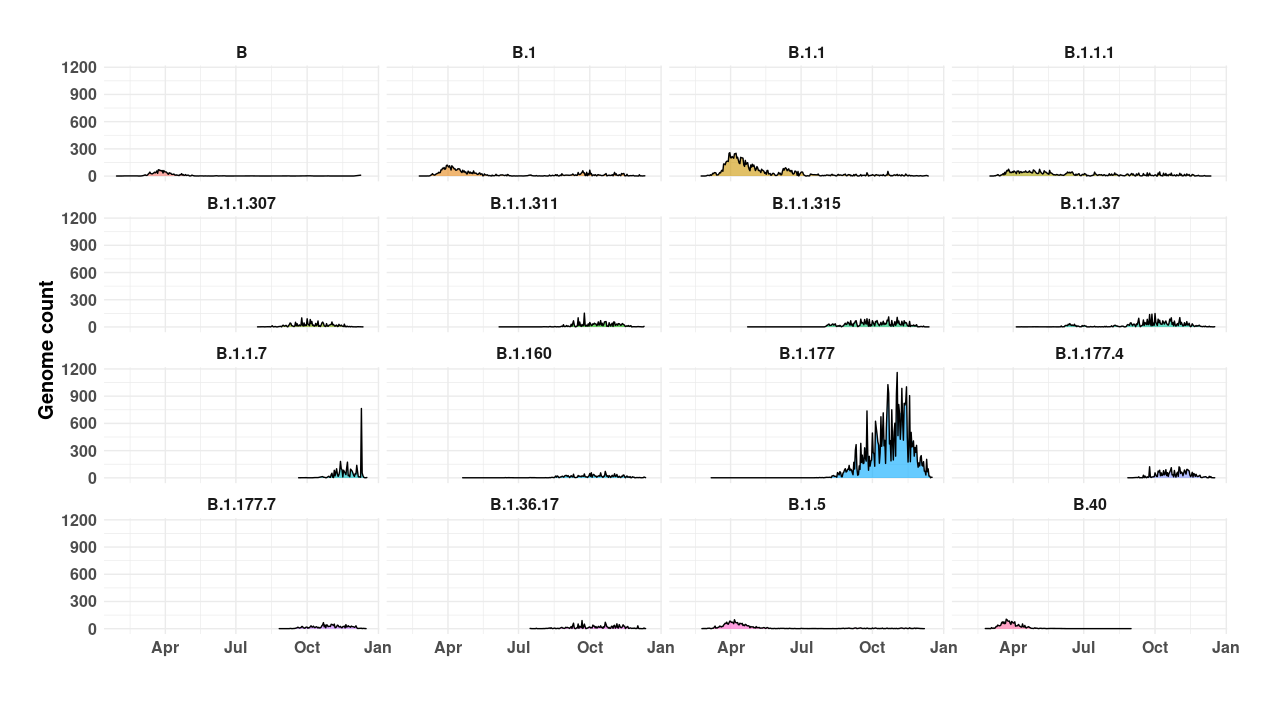
Cov-lineages has
a lot of good information on all of these lineages, including B.1.177 which
has recently set off alarms as the new “mutant UK strain”.
Code used to make the above plots can be viewed here.