Recently I was asked by a friend if they knew about any databases that classified cannabis strains by symptoms people tend to use them to relieve. I didn’t know of the existence of any but had heard about leafly.com which catalogues user reviews of various cannabis strains and compiles data on their characteristics.
I thought this could be a good place for them to start and so I started looking into what it would take to make a webscrapper to pull down all the data leafly has complied on hundreds on cannabis strains.
It turns out it didn’t take that much.
First, you can browse strains by going to the strains url at https://www.leafly.com/strains.
Conveniently, you can iterate through pages by just adding ?page=n where
n is whatever page you want.
I started looking through the html of these pages originally to find a way to pull out urls that would lead to each individual strain’s page but leafly (conveniently for me) provides basically all the information it has on the strains listed on a specific page in a nicely formatted json string in a script at the bottom of the page.
I was using the BeautifulSoup package and so after getting the page content
with requests all it took was
json_script = str(soup.find_all('script', id='__NEXT_DATA__'))
clean_json = re.sub('<.*?>', '', json_script)
json.loads(clean_json).pop()
to extract, clean and load the json into a dictionary. I did this for all pages and collected 167 json files.
I then threw together a quick python script to pull out basic attributes about each strain from the json file collection including metrics on the terpene content of each strain and the feelings strains tend to create for users, collectively called “effect measurements”. These were both expressed as floats but I am not sure what the units could be for either if there are any.
First I did some visualization in R to see if there were any differences between the phenotypes that immediately popped out. Below are two heatmaps comparing effect measurements and terpenes (right to left respectively). The color of the bar on the y-axis corresponds to the strains phenotype.
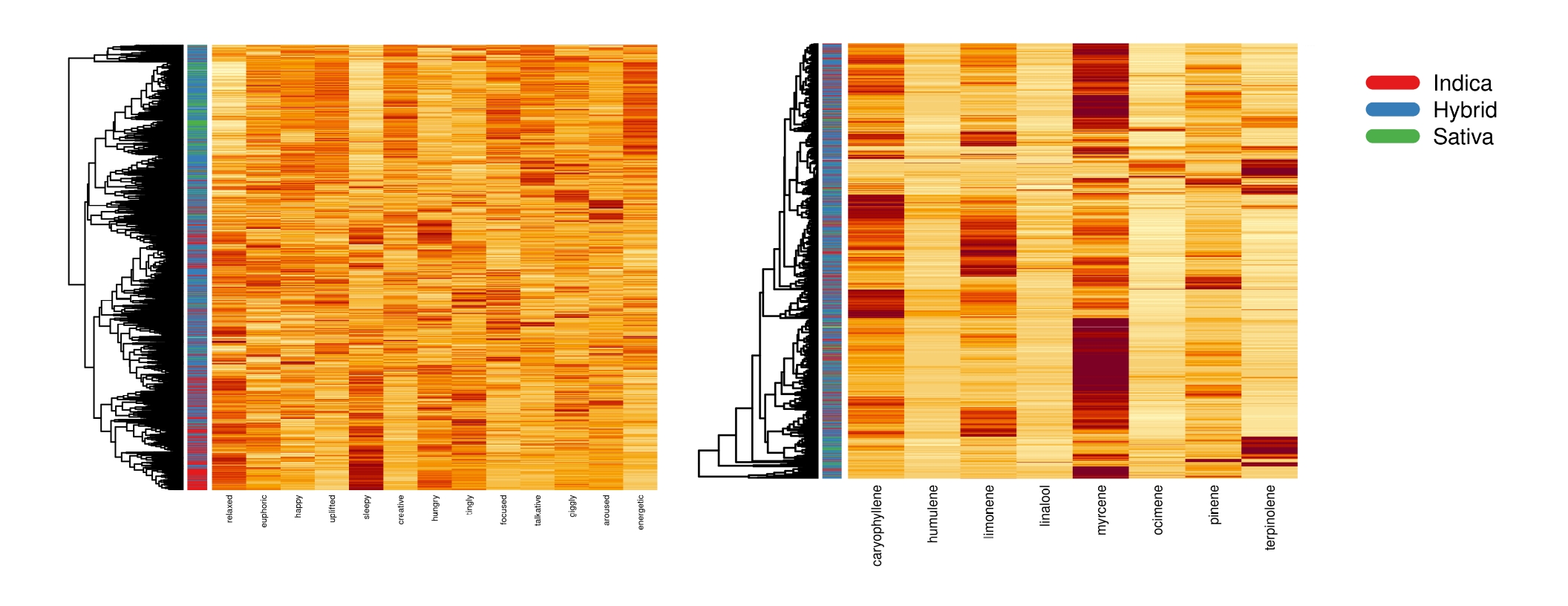
While there was not obvious clustering when using terpene measurements, there was slightly more promising results going off the effect measurements; this was actually opposite what I was expecting.
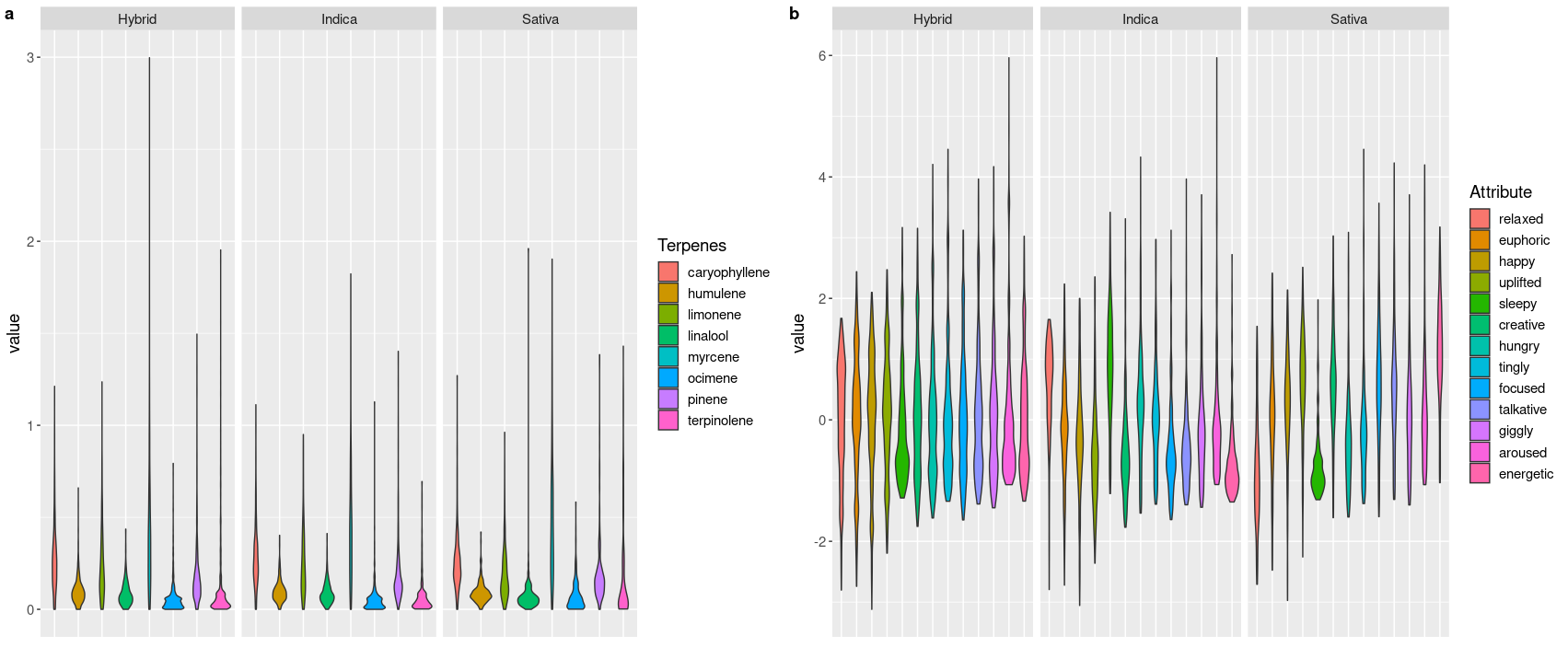
I also plotted the same attributes shown in the heatmaps as violin plots, separated by each phenotype which are shown below.
I wanted to see if I could use classify the phenotype of each strain (sativa, indicia or hybrid) based on the various terpene and / or effect measurements in a way that was at least better than a random guess.
I choose to go with a random forest model using the randomForest library in
R since there are three possible classifications. The variable importance
plots generated from the varImpPlot function after training the models on their
respective training sets is below.
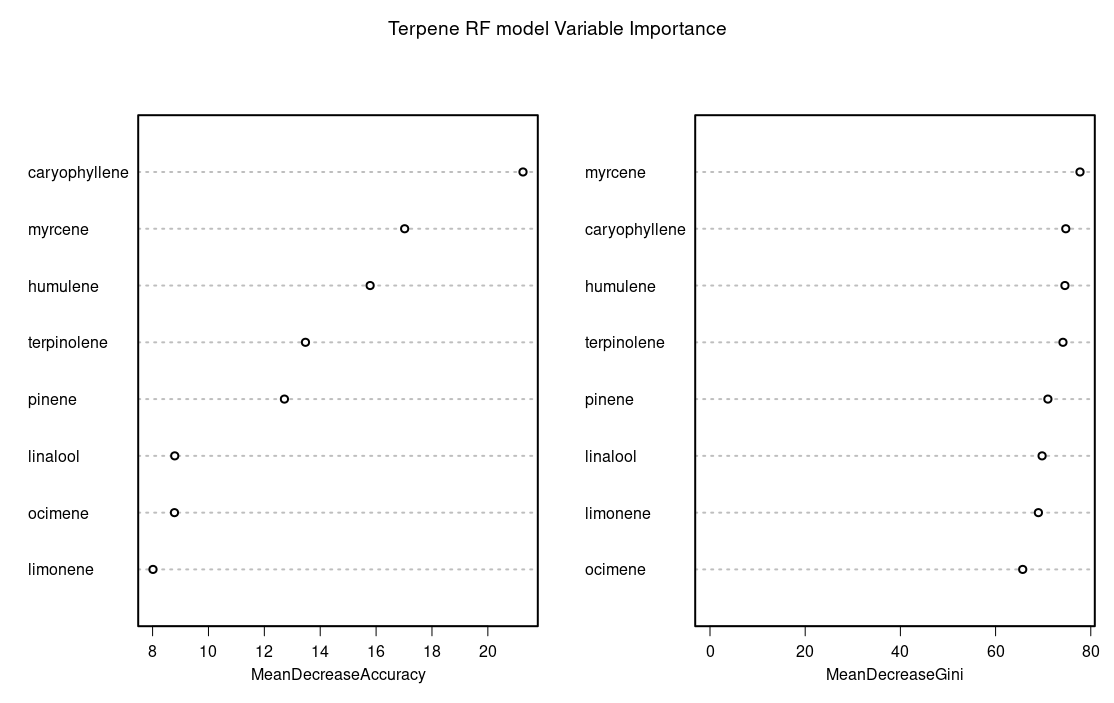
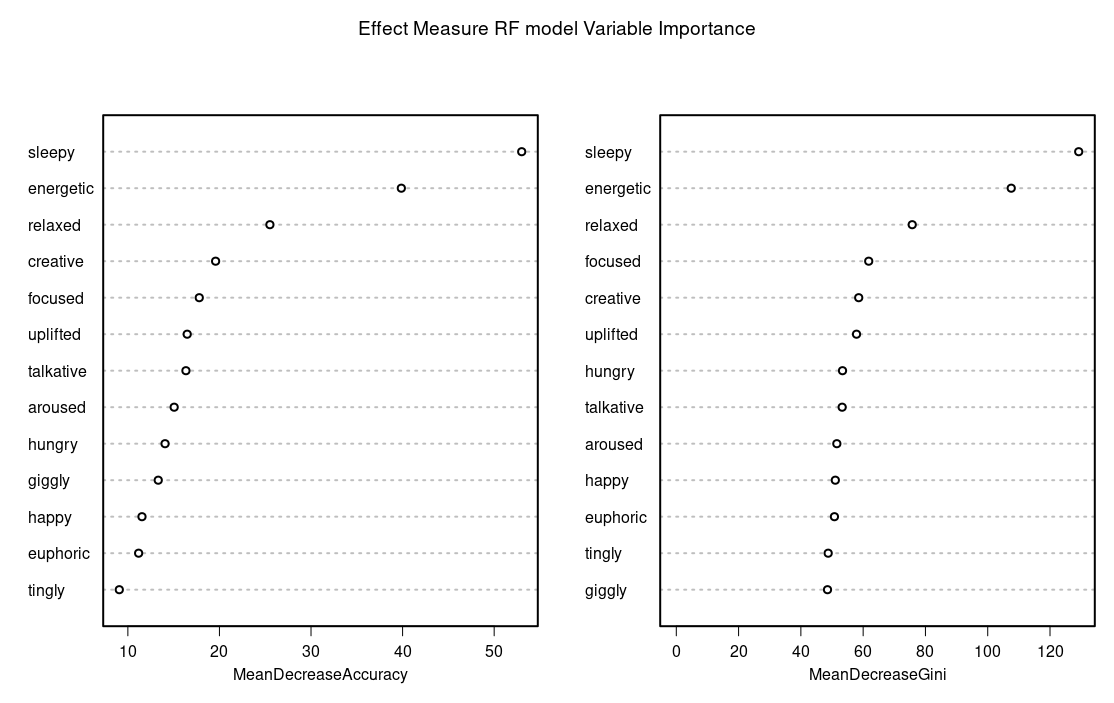
Which indicated sleepy as the most predictive measure of effect variable and caryophyllene or myrecene as the most predictive terpenes.
I then ran each model on their validation sets and created a confusion matrices.
Terpene confusion matrix
predicted
observed Hybrid Indica Sativa
Hybrid 685 25 10
Indica 243 8 0
Sativa 123 7 3
Using terpenes as the explanatory variables was worse then guessing and basically the model just thought everything was a hybrid. This could have been because most of the strains were hybrids.
Effect measurement confusion matrix
predicted
observed Hybrid Indica Sativa
Hybrid 845 56 43
Indica 180 213 1
Sativa 157 1 91
Not great either, but better. This could suggest that user ratings are slightly more predictive of strain phenotype then leafly terpene measurements. Although, there is a lot of optimization that could still be done here. Either way, fun little exercise that traversed web-scrapping, Python and R.
If you would like to play around with the processed data (csv) you can download it at this link.